Nos développements méthodologiques RMN pour l’analyse quantitative de mélanges complexes répondent aux besoins d’une large communauté d’utilisateurs en métabolomique et fluxomique.
L'équipe explore particulièrement le potentiel d’approches non-conventionnelles qui peuvent s’avérer hautement complémentaires aux méthodes 1D conventionnelles, aussi bien pour le profilage non ciblé que pour l’analyse quantitative ciblée de mélanges biologiques complexes. Ces méthodes comprennent la RMN 2D ultrarapide à haut et bas champ magnétique, la RMN hyperpolarisée ainsi qu’une approche originale «métabisotopomique» combinant la métabolomique et l’analyse isotopique.
Étant spécialisés en méthodologie, l'équipe ne cible pas d’application spécifique, préférant explorer une large gamme de questions de recherche en lien avec nos collaborateurs en agroalimentaire, sécurité chimique des aliments, médecine légale, santé ou sciences végétales. Notre équipe fait partie de l’infrastructure de recherche MetaboHUB qui regroupe les centres d’expertise français en métabolomique et fluxomique, et de la plateforme métabolomique Corsaire (Biogenouest).
Current team members: Jérémy Marchand, Aurore Michaud, Estelle Martineau, Jonathan Farjon, Serge Akoka, Marine Letertre, Patrick Giraudeau*
Past members: Adrien le Guennec
Collaborations (past and present): Catherine Deborde and Annick Moing (INRAe Bordeaux), Gaud Dervilly-Pinel and Yann Guitton (LABERCA Nantes), Olivier Grovel and Samuel Bertrand (MMS Nantes), Jean-Charles Portais (INSA Toulouse), Pascal de Tullio (Univ. Liège), Cécile Canlet (Axiom Toulouse), Mikaël Croyal (M-SHARK Nantes), Julien Boccard (Université de Genève)
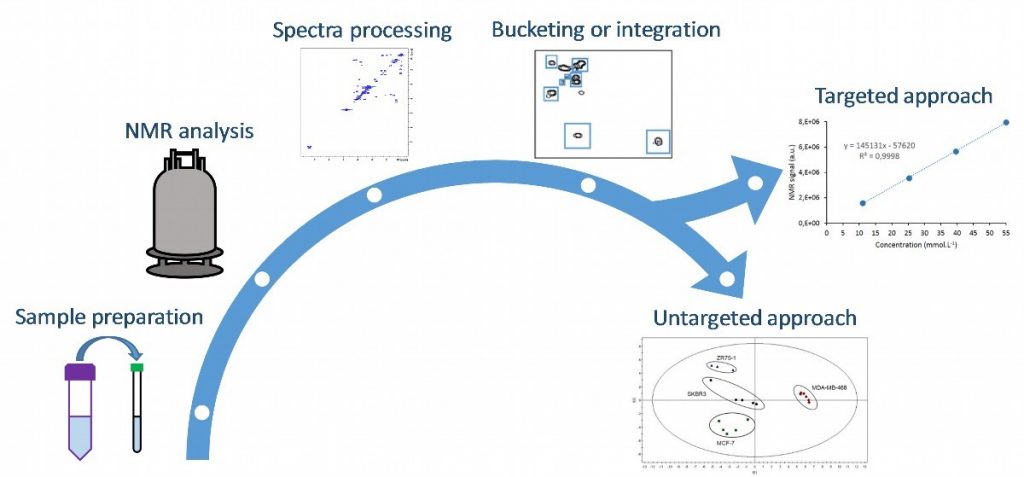
While NMR-based metabolomics studies are mostly performed with 1H 1D NMR, the latter suffers from the severe and numerous peak overlaps characterizing complex biological samples. In this context, we are evaluating the potential of various 2D NMR approaches for a variety of targeted and untargeted metabolomics applications. Fast and repeatable 2D NMR methods are indispensable in this field, not only for the sake of high-throughput analysis, but also to reduce the impact of hardware instabilities and consequently improve the precision.
Multi-dimensional methods based on ultrafast 2D NMR or non-uniform sampling have already been applied in different fields through past and current collaborations. These applications include the targeted quantification of metabolites in extracts, or the untargeted metabolomics or lipidomics analysis of various samples, to address research questions in chemical food safety, chemical ecology, or health. This strategy is explored both at high field and at medium field (benchtop spectrometers). The continuum of fast 2D NMR methods for metabolomics and lipidomics studies are also being explored by putting these methods in competition through multiblock integration. The results prove the promises of fast 2D NMR methods to facilitate the bucketing step and thus increase statistical analyses performances.
References:
- B. Gouilleux, J. Marchand, B. Charrier, G. S. Remaud, P. Giraudeau, High-throughput authentication of edible oils with benchtop Ultrafast 2D NMR, Food Chemistry, 244, 153 (2018)
- A. Marchand, E. Martineau, Y. Guitton, B. Le Bizec, G. Dervilly-Pinel, P. Giraudeau, A multidimensional 1H NMR lipidomics workflow to address chemical food safety issues, Metabolomics 14, 60, (2018)
- P. Giraudeau, NMR-based metabolomics and fluxomics: developments and future prospects, Analyst, 145, 2457, (2020).
- B. Féraud, E. Martineau, J. Leenders, B. Govaerts, P. de Tullio, P. Giraudeau, Metabolomics, Combining rapid 2D NMR experiments with novel pre-processing workflows and MIC quality measures for metabolomics, Metabolomics, 16, 42 (2020)
- E. Martineau, J.-N. Dumez, P. Giraudeau, Fast quantitative 2D NMR for metabolomics and lipidomics: A tutorial, Magn. Reson. Chem.58, 390 (2020).
- M. Letertre, P. Giraudeau, P. De Tullio, Nuclear magnetic resonance spectroscopy in clinical metabolomics and personalized medicine: current challenges and perspectives, Frontiers in Molecular Biosciences 8, 698337 (2021)
- C. Praud, M. Letertre, A. Dey, J.N. Dumez, P. Giraudeau, Fast 2D NMR for Metabolomics, Fast 2D Solution-state NMR: concepts and Applications, 377-414 (2023)
Supported by: ANR (MetaboHub), Région Pays de la Loire (RFI Food 4.2 Lipidotool), Biogenouest (CORSAIRE platform)
Current team members: Benoît Charrier, Patrick Giraudeau, Jonathan Farjon*
Collaborations : Olivier Gonçalves, (GEPEA, Nantes Univ.)
Past team members : Dylan Bouillaud, Jérémy Pruvost and Jack Legrand
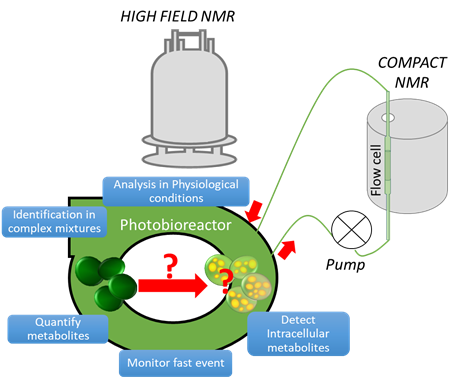
The first microorganisms we investigate by NMR were microalgae as photosynthetic organisms with a very high biochemical diversity and a great ability to produce valuable biomass. Only a minor part of the millions of species living on earth are currently known, therefore microalgae form a subject with a strong investigation potential. Some microalgae such as Parachlorella Kessleri and Nannochloropsis Gaditana are known to accumulate lipids under nitrogen starving stress conditions. This behavior is interesting because it forms the basis of the third-generation biofuel production, a promising alternative energy.
In this context, we explored the potential of multiscale NMR for the analysis of microalgae cells and their lipidic extracts. The main challenge was to identify and quantify lipids in microalgae aqueous cultures. With this aim, high field NMR provides the best sensitivity and resolution to reveal lipids from extracts. Moreover, compact flow NMR was a suitable apparatus for real time online monitoring of bioprocesses such as the accumulation of lipids in microalgae during a nitrogen starvation in photobioreactor.
In addition to instrumental improvements for respecting physiological conditions by flowing cultivations, NMR pulse sequences development, especially in solvent suppression, ultra-resolved and fast NMR are very promising for tracking the metabolism of microalgae.
We are starting a new project on the control of Phaeodactylum Tricornutum metabolism able to exudate lipids droplets in the cultivation medium for making energy. Moreover, in the field of space missions in limited room area, another project with the European Space Agency will be dedicated to show performances of compact flow NMR to control Spirulina to produce highly nutritive metabolites as food for astronauts.
References:
- D. Bouillaud, J. Farjon, O. Gonçalves, P. Giraudeau, Magnetic Resonance in Chemistry 2019, 57, 794–804.
- T. Castaing-Cordier, D. Bouillaud, P. Bowyer, O. Gonçalves, P. Giraudeau, J. Farjon, ChemPhysChem 2019, 20, 736–744.
- D. Bouillaud, V. Heredia, T. Castaing-Cordier, D. Drouin, B. Charrier, O. Gonçalves, J. Farjon, P. Giraudeau, Algal Research 2019, 43, 101624.
- D. Bouillaud, D. Drouin, B. Charrier, C. Jacquemmoz, J. Farjon, P. Giraudeau, O. Gonçalves, Process Biochemistry 2020, 93, 63–68.
Supported by Région Pays de la Loire (Pari scientifique AMER-METAL 2017-2020), CNRS (Projet interdisciplinarité RMN-(ME)2-TAL 2017-2020), European Space Agency (SPACE_ALG 2024-2027), PEPR B-BEST (ALGADVANCE 2023 - 2028)
Current team members: Tania Mhanna, Margot Sanchez, Virginie Silvestre, Mathilde Grand, Patrick Giraudeau, Serge Akoka*, Gérald Remaud.
Past team members: Lenny Haddad, Vincent Portaluri, Ghina Hajjar, Denis Loquet, Boris Gouyeux, Sophie Guyader, Noëlle Merchack. Collaborations: J. Bejjani, T. Rizk (Saint-Joseph University, Beirut, Lebanon), F. Thomas, E. Jamin (EUROFINS, Nantes, France).
We apply high-field NMR combined with advanced statistical analysis methods for the profiling of complex samples (biological fluids, extracts, foodstuffs, etc.). This profiling approach forms part of our “omics” expertise as a member of the CORSAIRE metabolomics core facility (part of the Biogenouest network). Profiling tools include 1D 1H and 13C NMR, but also innovative fast 2D NMR methods. These approaches are applied both at high-field and on bencthop NMR spectrometers.
Beyond the classical ”omics” workflow, particularly original work concerns the development of profiling methods for lipid mixtures as found in edible fats. Such mixtures may be authenticated by the metabolic profile but also by the determination of the isotopic profile of major components. We show that a combined 13C-isotopomics and metabolomics approach presents many advantages: no need for prior chemical manipulations, determination of the positional distribution of major fatty acids on the glycerol backbone of triglycerides, and measurement of the relative position-specific isotopic 13C content at different chemical sites.
Furthermore, all this information is obtained from the same spectrum in a few minutes. This method has been compared with 1H profiling and gas chromatography. Its higher efficiency is explained by a significant contribution of variables that can only be obtained by this technique. Generally speaking, 13C profiling enables the classification of all types of edible fats.
References:
- N. Merchak, T. Rizk, V. Silvestre, G.S. Remaud, J. Bejjani, S. Akoka. Olive oil characterization and classification by 13C-NMR with a polarization transfer technique: A comparison with gas chromatography and 1H-NMR. Food Chemistry, 2017, 245, 717-723.
- S. Guyader, F. Thomas, V. Portaluri, E. Jamin, S. Akoka, V. Silvestre, GS Remaud. Authentication of edible fats and oils by non-targeted C-13 INEPT NMR spectroscopy. Food Control 91, 216-224 (2018).
- G. Hajjar, T. Rizk, S. Akoka, J. Bejjani. Cholesterol, a powerful 13C isotopic biomarker, Analytica Chimica Acta. 1089, 115–122 (2019).
- G. Hajjar, N. Merchak, C. Daniel, T. Rizk, S. Akoka, J. Bejjani. Improved Lipid Mixtures Profiling by 1H NMR Using Reference Lineshape Adjustment and Deconvolution Techniques. Talenta, 208 (2020).
- G. Hajjar, L. Haddad, T. Rizk, S. Akoka, J. Bejjani. High-resolution 1H NMR profiling of triacylglycerols as a tool for authentication of food from animal origin: Application to hen egg matrix. Food Chemistry 320 130056 (2021).
Supported by: National Council for Scientific research of Lebanon, Saint-Joseph University of Beirut, EUROFINS Nantes, National Council for Scientific research and University of Nantes.
Current team members: Virginie Silvestre, Benoît Charrier, Victor Ribay, Patrick Giraudeau, Marine Letertre*
Collaborations (past and present): Gaud Dervilly, Yann Guitton and Anne-Lise Royer (LABERCA Nantes), Daniel Jacob (INRAE Bordeaux), Cécile Canlet (Axiom Toulouse), Mikaël Croyal and Chloé Cloteau (M-SHARK Nantes), Julien Boccard (Geneva University)
We are currently exploring the applicability of the non-conventional NMR methods developed in the team (fast 2D, hyperpolarised and benchtop NMR) to pre-clinical metabolomics to overcome the current limitations of current 1H 1D NMR-based metabolomics. These methods that are more sensitive, highly resolved and/or more accessible, taken in combination with the advantages of Mass Spectrometry (MS), have the potential to strengthen personalized medicine. Fast 2D NMR methods allow to spread signals over two dimensions and have thus proven to facilitate metabolite identification6–8. Hyperpolarized NMR provides a major sensitivity enhancement9 and dissolution-dynamic nuclear polarization (d-DNP) coupled to NMR was successfully applied for the first time to urine samples10. Fast 2D NMR methods were successfully applied to various samples including pig plasma extracts and d-DNP was shown to be highly promising for the analysis of urine samples.
Furthermore, we are gaining in competences in the targeted profiling of pre-clinical biomarkers for specifics pathologies. For instance, 1D 1H lipoprotein NMR profiles were recorded on plasma from patients with type 2 diabetes administrated with either placebo or alirocumab, provided key results showing that NMR (1) gives absolute concentrations which highly correlate with the ones obtained by other clinical analysis (Fig. a) and (2) gives access to biomarkers, such as LDL-triglycerides and -ApoB100 (among many others lipoproteins-related biomarkers), which are very complicated to obtain by MS or clinical tests without numerous and tedious sample manipulations (Fig. b).
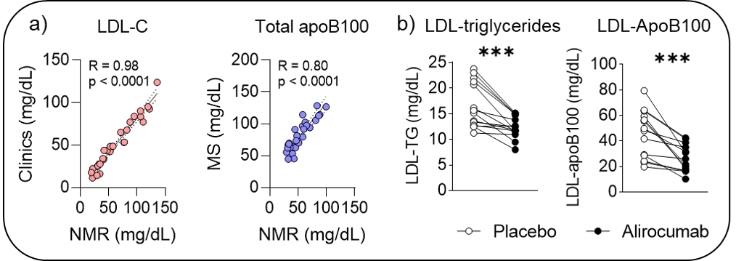
a) Correlations between LDL-Cholesterol and apoB100 NMR concentrations with clinical assay and ones, respectively, and b) NMR LDL-triglycerides or LDL-ApoB100 concentrations.
Key References:
- M. Letertre, P. Giraudeau, P. De Tullio, Nuclear magnetic resonance spectroscopy in clinical metabolomics and personalized medicine: current challenges and perspectives, Frontiers in Molecular Biosciences 8, 698337 (2021)
- V. Ribay, A. Dey, B. Charrier, C. Praud, J. Mandral, J.N. Dumez, M. Letertre, P. Giraudeau, Hyperpolarized 13C NMR Spectroscopy of Urine Samples at Natural Abundance by Quantitative Dissolution Dynamic Nuclear Polarization, Angewandte Chemie International Edition 62 (27), e202302110 (2023)
- V. Ribay, C. Praud, M. Letertre, J.N. Dumez, P. Giraudeau, Hyperpolarized NMR metabolomics, Current Opinion in Chemical Biology 74, 102307 (2023)
Supported by: ANR (MetaboHub), Biogenouest (CORSAIRE platform), and European Research Council (CoG SUMMIT)
Current team members: Patrick Giraudeau, Marine Letertre*
Collaborations (past and present): Gaud Dervilly, Yann Guitton and Anne-Lise Royer (LABERCA Nantes), Mikaël Croyal and Chloé Cloteau (M-SHARK Nantes), Julien Boccard (Geneva University)
To benchmark the newly proposed methods described in the ‘NMR-based metabolomics’ section, between each other but also to the other analytical foundations of the clinical metabolomics community (MS)ultiblock integration. This approach aims at putting these methods in competition, to capture a larger part of biomolecular pathways and to improve biological interpretation11.
Encouraging results were obtained for the use of fast 2D NMR methods in metabolomics by analyzing plasma extracts from pigs exposed or not to bisphenol A (BPA) and analyzed by 1D NMR and three fast 2D NMR methods. The latter are those which capture the largest part of the inter-sample variability, probably thanks to better resolved buckets compared to 1D NMR (Fig.).
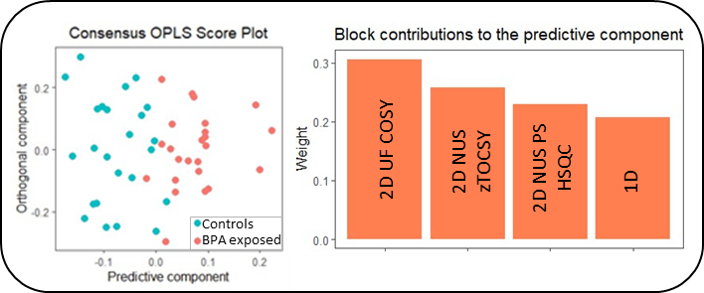
Key References:
- M. Letertre, G. Dervilly, P. Giraudeau, Combined nuclear magnetic resonance spectroscopy and mass spectremtry approaches for metabolomics, Analytical Chemistry 93, 1, 500-518 (2020)
Supported by: ANR (MetaboHub), Biogenouest (CORSAIRE platform), and European Research Council (CoG SUMMIT)
